
What is Spatial Transcriptomics?
Spatial relationships between cells and structures are critical to their development and pathophysiology. With the emergence of spatial transcriptomics technologies, we can now bridge histology and genomics to deepen our understanding of cellular organization and gene expression within a tissue of interest.
With our 10X Genomics Visium CytAssist workflow, the whole transcriptome is mapped within the histological context of the tissue using either formalin fixed paraffin embedded (FFPE) blocks or pre-sectioned tissues on glass slides as starting material, giving you the ultimate flexibility.
We provide a complete seamless package of end-to-end wet lab to dry lab services meaning we can take care of everything from sample processing to data analysis enabling you to gain valuable, reproducible, high-quality insights. All we need are formalin fixed paraffin embedded (FFPE) tissue blocks or tissue sections (stained or unstained) on glass slides and we will take care of the rest.

Our Visium CytAssist Process
Key Benefits of Spatial Transcriptomics at AcelaBio
At the forefront of science and innovation, AcelaBio is one of the first laboratories to offer a complete end-to-end spatial transcriptomics workflow leveraging 10X Genomics Visium CytAssist. Our seamless, high-quality process from wet lab to dry lab translates your samples into meaningful biomarker insights.
Unlock the potential of your tissue sample
The key to the 10X Visium Spatial Gene Expression solution is the Visium CytAssist Spatial Slide with capture areas of 6.5 x 6.5 mm or 11 x 11 mm. For 6.5 mm2, each of its two capture areas includes ~5,000 barcoded spots, for 11 mm2, each of its two capture areas includes ~14,000 barcoded spots, which in turn contain millions of spatially barcoded capture oligonucleotides. Upon permeabilization, tissue mRNA is released and attaches to the barcoded oligos, enabling the collection of gene expression data.
From early-stage discovery to clinical research, spatial transcriptomics can unravel novel insights and accelerate biomarker discovery within the histological context.

We Make Spatial Transcriptomics Look Easy
Define and visualize tissue architectures from spatially resolved transcriptomics with AcelaBio.
Our experienced scientific team provides seamless delivery from project initiation to delivery of data.

Deriving Novel Insights to Accelerate Your Research
Spatial transcriptomics generates ‘big’ data requiring programming and statistical expertise to maximize biomarker discovery
We are serious about data analysis
Our analysis and visualization packages go beyond the standard assessments and dive deep into the data to increase the value of your output. We offer a flexible, iterative three-step approach based on your study objectives with the goal of helping you understand what your data means through integration with highly curated datasets.

Quality is key. Our first step is to pre-process the data and evaluate quality metrics to ensure the data meets our quality standards and is ready for analysis in our downstream bioinformatics pipelines. Sequencing data will be aligned to the appropriate reference transcriptome, mapped to the histopathology image and gene counts will be determined using the 10X Genomics recommended Space Ranger pipeline.
Key Deliverables:
- Tissue positions
- Scale factors
- Image outputs
- Feature-Barcode Matrices
- QC report

To explore the spatial gene expression data, our analysis pipeline is designed to classify the data into clusters and annotated histological features, explore cluster proportions per experimental condition, and find spatially-resolved biomarkers.
Key Deliverables:
- Clustering UMAP and Spatial graphs
- Cluster proportions graph and table
- Gene markers heatmap and table
- Gene markers spatial heatmap
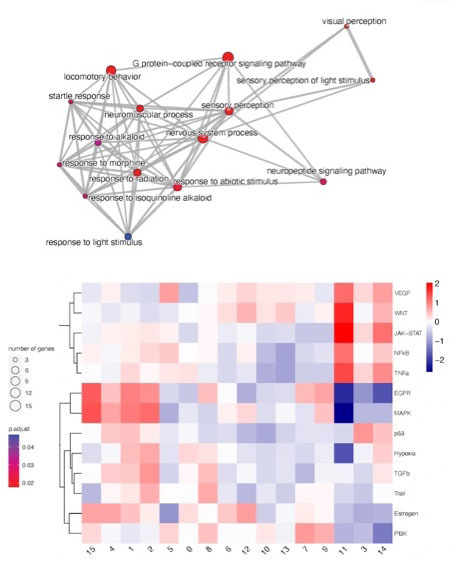
Once you know what genes are active and where the activity is located in the tissue, the key question is, what does your data mean? We provide context to your results by integrating your data with high-quality curated datasets of biomarkers, cell types and signaling pathways to better understand the cellular heterogeneity, annotate cell subtypes, identify significantly enriched signaling pathways, and map pathway activity into the histology slides. As we unravel novel insights, we will work iteratively with you to achieve the depth of analysis that meets your needs.
Key Deliverables:
- Identification of enriched signaling pathways by Gene Set Enrichment Analysis (GSEA)
- Mapping pathway activity into the histology slides
- Identification of cell composition

Partner with a 10X Certified Service Provider
As one of the earliest adopters of 10X Genomics’ Visium CytAssist technology, we pride ourselves on being a trusted spatial transcriptomics partner to leading pharmaceutical and biotechnology companies. AcelaBio is a 10X Certified Provider and has been recognized by 10X Genomics for its expert ability to deliver high quality, reproducible spatial transcriptomics data in a timely manner.
What tissue types do we work with?
When submitting FFPE tissue, the Visium Spatial Gene Expression solution is available for human and mouse tissue samples. We can work with blocks, or tissue sections affixed to glass slides (stained or unstained). For the complete list of tested tissue, please refer to the 10X Genomics website.
What is the size of the spots on the Visium Spatial Slide?
Spots are 55 µm in diameter.
Does Visium enable single cell resolution?
p class=”mb-4″>The current Visium solution has barcoded spots that are 55 µm in diameter which captures approximately 10 cells. Through our bioinformatics approach, we can perform data deconvolution to analyze data at the cellular level.
What is the configuration of the capture area of the Visium Spatial Slide?
The capture area is 6.5mm x 6.5mm or 11mm x 11mm.






